Research
Single cell
Deciphering lineage decisions in the human thymus at the single cell level
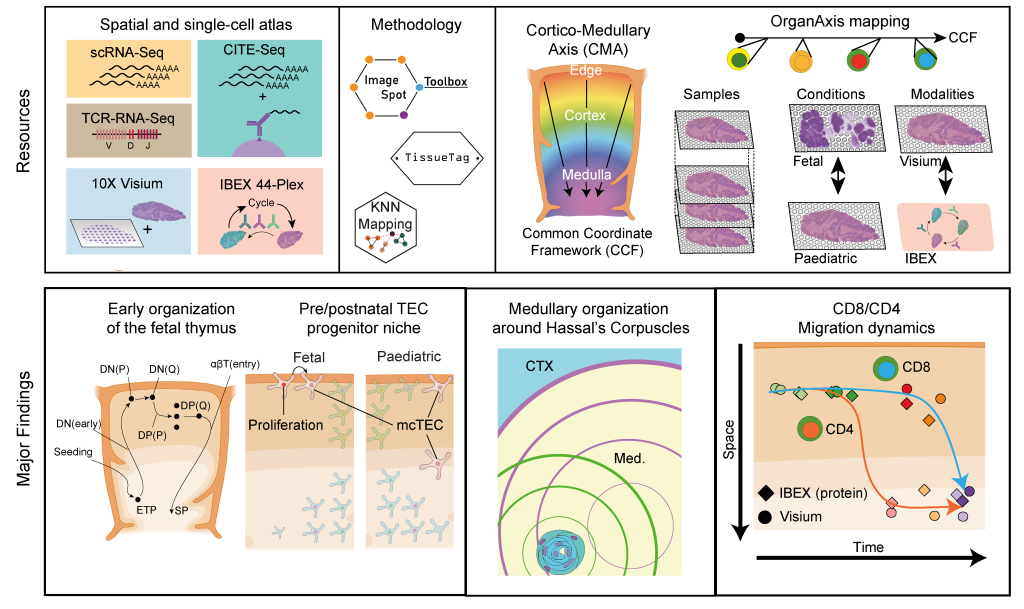
In recent years, advances in single cell sequencing technologies have aided the analysis of molecular processes, such as changes in transcription and chromatin landscape, at an unprecedented resolution. This has been groundbreaking for the identification and characterization of rare cell types as well as the exploration of continuous differentiation trajectories, which cannot be achieved using bulk sequencing approaches. In our lab, we are using a variety of droplet-based single cell techniques to study the driving forces behind critical lineage decisions and cell differentiation in human thymocytes. In this context, we have been involved in the Human Cell Atlas initiative and have generated a cellular atlas of the human thymus, which helped map both fetal and pediatric thymopoiesis at single cell resolution1. More recently, through a collaboration with the labs of Sarah Teichmann (Wellcome Sanger Institute) and Ron Germain (NIAID, NIH) we have investigated this process from a spatial perspective by employing spatial transcriptomics and multiplex protein imaging. Through the development of a common coordinate framework for the human thymus we were able to map the localization of T cell precursors, stromal cells and other thymic cell types throughout early development. This revealed that, despite ongoing changes in the macroscopic tissue architecture, the thymocyte maturation trajectory is already established in the first trimester of fetal development. Moreover, using CITE-seq to achieve a detailed annotation of thymocyte differentiation stages, we discovered distinct maturation kinetics between CD4 and CD8 lineage thymocytes upon their positive selection.2
Unravelling the molecular mechanisms controlling the differentiation of thymus seeding precursors
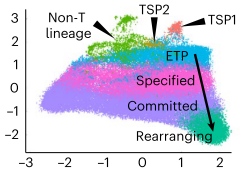
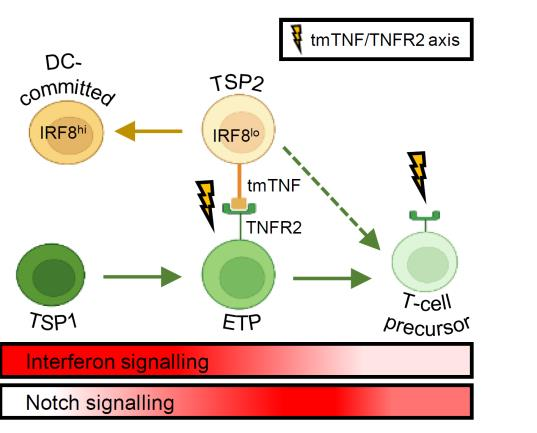
The thymus is continuously colonized by bone-marrow derived hematopoietic precursor cells and the local microenvironment gradually converts these multipotent thymus seeding progenitors (TSPs) to become T lineage-committed. We have recently identified two distinct TSP populations using scRNA-seq data from immature thymocytes, bone marrow and peripheral blood3. In silico fate predictions suggested that one subset (TSP1) resembles canonical T cell precursors, whereas the other subtype (TSP2) may also give rise to dendritic cells (DCs). In a subsequent study we were indeed able to validate in vitro that T cells and DCs can arise from a common thymic precursor, which resembles the TSP2 population4. Moreover, we found that DC-primed thymocytes were able to support the differentiation of progenitor cells along the T lineage via membrane-bound TNF, confirming the involvement of hematopoietic stromal cells in the early stages of human T cell development. We aim to further characterize the various precursor populations present in the human postnatal thymus and determine the molecular processes governing the commitment to the T and possibly other hematopoietic lineages.
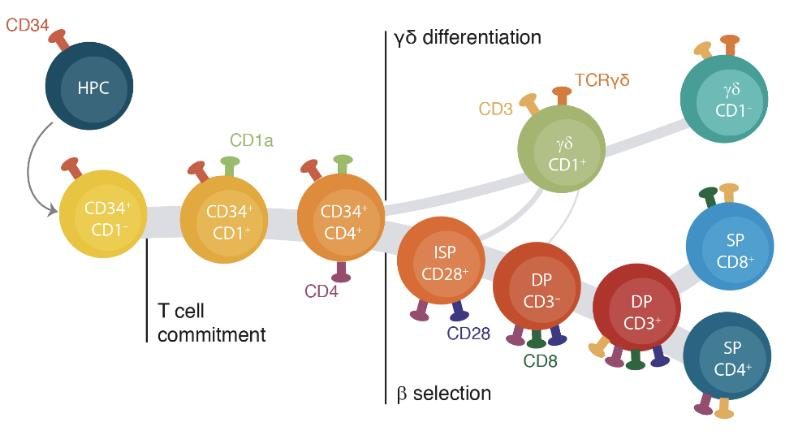
Identifying the drivers of the αβ vs. γδ lineage decision in human thymocytes
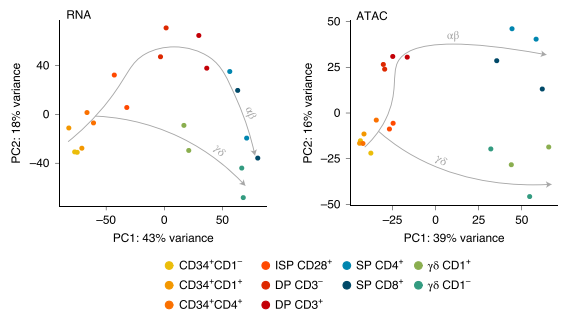
αβ and γδ T cells have distinct roles in the immune system but arise from common precursors in the thymus. Although the lineage bifurcation has been studied extensively in mice, critical differences have been described between the two species5 and our understanding of the molecular mechanisms involved in this developmental process in the human context remains vague. Our lab has employed bulk sequencing methods to study gene expression and epigenetic characteristics in 12 discrete differentiation stages of human thymocytes, including bipotent precursors and immature/mature cells of the αβ- and γδ-lineage6. This revealed several disparities between the two lineages, such as differences in chromatin remodeling and abundance of certain histone marks. To overcome the constraints of bulk technologies we are aiming to employ multi-modal single cell approaches on primary human thymocytes to determine the drivers of these lineage-specific changes and their role in the αβ vs. γδ lineage decision process.
iPSC
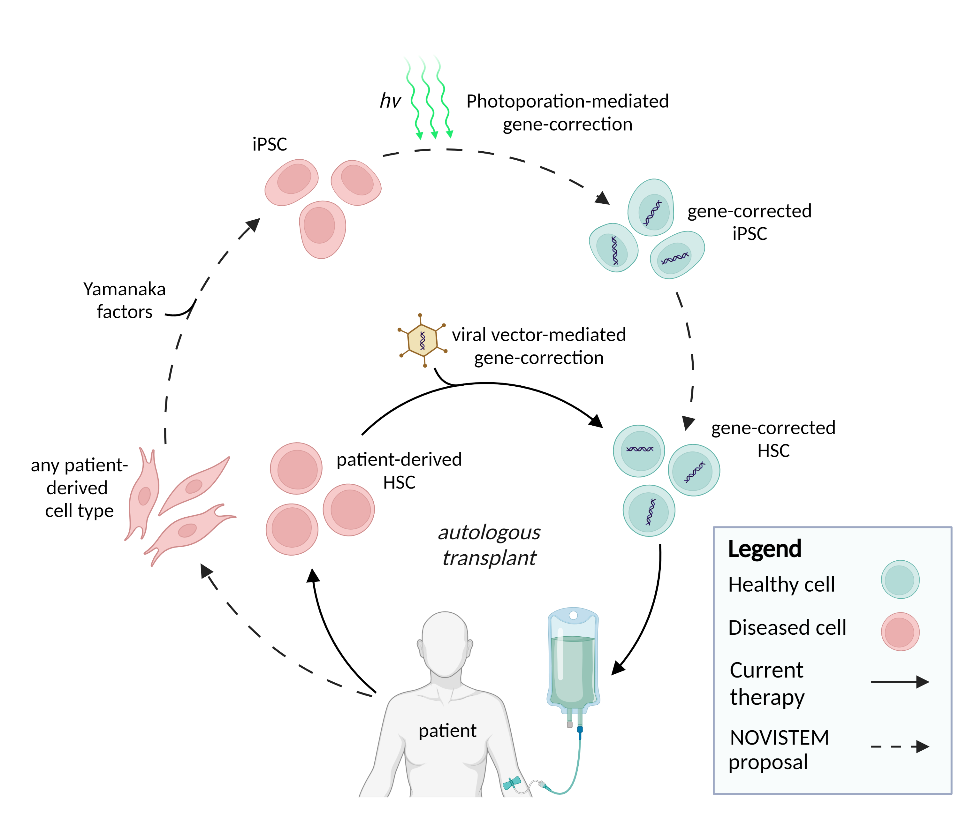
Investigating the potential of induced pluripotent stem cells as a novel source for generating cell therapy products
Despite the increasing awareness that cell and gene-therapy approaches have tremendous biomedical potential, their broad clinical application has been challenging due to prolonged and expensive production times and the emergence of severe immune- and gene-delivery dependent side effects. Therefore, one of our research aims is to establish a stream-lined and high-throughput protocol for the generation of off-the-shelf cell therapy by combining the photoporation platform of Prof. Kevin Braeckmans (Faculty of Pharmaceutical Sciences) for gene delivery with a breakthrough biological concept that will permit to manufacture functional, gene-corrected blood forming stem cells and chimeric antigen receptor (CAR) T cells that can recognize and kill specific tumor cells through this CAR. To achieve this, we will use and optimize photoporation for CRISPR-mediated and site-specific gene-editing to obtain controlled CAR expression and for performing gene-correction in induced pluripotent stem cells (iPSCs). These are stem cells that have the intrinsic potential to generate any cell type of the body but that are derived from fully differentiated cells following reprogramming. From these gene-modified iPSCs, we wish to generate CAR T cells and blood forming stem cells, respectively, by selectively targeting a signaling pathway that we established to be critical in human blood cell development and T cell development. Following functional validation of the generated cell products, we aim to optimize the current protocols to increase the potential for clinical implementation and establish a high-throughput platform to generate a large number of CAR expressing iPSC lines from different ages, sex and ethnicities to demonstrate the population-wide implementation potential of our approach. This will allow to generate a bank of well-characterized, CAR expressing iPSC that can be used as of-the-shelf cell therapy products in the absence of any rejection-related issues, thereby significantly advancing the currently implemented adaptive CAR T cell approaches by reducing the production costs and time, by selectively targeting the CAR into a well-controlled location which will prevent variability and by facilitating the production and evaluation of novel CARs for other cancer entities such as solid tumors. More information on this EU-funded research project can be found on the dedicated website: novistem.ugent.be.
GATA3
Unravelling the role of GATA3 in normal and malignant human T cell development
The transcription factor GATA3 is an important regulator of various normal developmental processes and mutations in this gene have been described in a variety of cancers, including leukemia. During normal blood cell development, it has been shown in mice that GATA3 is essential for T cell development, not only as a positive regulator of this particular blood cell lineage, but also as a repressor of alternative lineage differentiation, such as B cell development. Within the context of leukemia, GATA3 mutations have been described in early T cell precursor acute lymphoblastic leukemia (ETP-ALL). This type of leukemia is characterized by a mixed myeloid/T-lymphoid phenotype. Previously, our lab has shown that GATA3 is important during the early stages of human T cell development to induce T-lineage commitment at the expense of NK cell development.
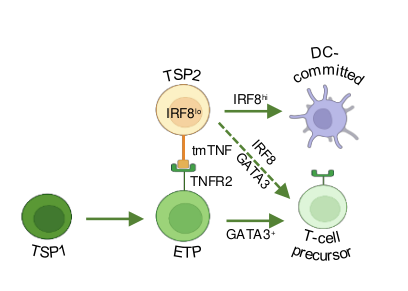
Given that these GATA3 mutations are considered to be loss of function mutations, we hypothesize that this phenotype might be caused by perturbations in the capacity of GATA3 to repress myeloid development, in addition to a developmental arrest in early T-lineage differentiation. Indeed, we recently also showed that GATA3 is required to repress IRF8 in TSPs to induce T-lineage differentiation at the expense of dendritic cell differentiation, suggesting that normal GATA3 function is critical to repress alternative lineage cell fate. Given that GATA3 also downmodulates Notch signaling, one of the most common drivers of T-ALL, we will also investigate how GATA3 mutations affect Notch activation in ETPs. Overall, we aim to unravel how GATA3 mutations are contributing to the emergence and maintenance of ETP-ALL. We anticipate that this will lead to the discovery of novel opportunities for therapeutic intervention.
References
1. Park, J. E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80-. ). 367, (2020).
2. Yayon, N., Kedlian, V.R., Boehme, L. et al. A spatial human thymus cell atlas mapped to a continuous tissue axis. Nature 635, 708–718 (2024). https://doi.org/10.1038/s41586-024-07944-6.
3. Lavaert, M. et al. Integrated scRNA-Seq Identifies Human Postnatal Thymus Seeding Progenitors and Regulatory Dynamics of Differentiating Immature Thymocytes. Immunity 52, 1088-1104.e6 (2020).
4. Liang, K. L. et al. Intrathymic dendritic cell-biased precursors promote human T cell lineage specification through IRF8-driven transmembrane TNF. Nat. Immunol. (2023) doi:10.1038/s41590-022-01417-6.
5. Boehme, L., Roels, J. & Taghon, T. Development of γδ T cells in the thymus – A human perspective. Semin. Immunol. 61–64, 101662 (2022).
6. Roels, J. et al. Distinct and temporary-restricted epigenetic mechanisms regulate human αβ and γδ T cell development. Nat. Immunol. 21, 1280–1292 (2020).
